Surface of a micelle¶
Summary¶
We use the GITIM class to investigate the surface composition of a DPC micelle.
This tutorial shows how to do some basic statistical analysis and how to visualize surface atoms / pockets for water.
The DPC micelle¶
The micelle example file MICELLE_PDB consists of 65 DPC and 6305 water molecules,
for a total of about 20,000 atoms (D. P. Tieleman, D. van der Spoel, H.J.C.
Berendsen, J. Phys. Chem. B 104, pp. 6380-6388, 2000)

As this is not a planar interface, we will use the GITIM class to identify the atoms at the
surface of the micelle. Our reference group for the surface analysis includes therefore
all DPC molecules
>>> import numpy as np
>>> import MDAnalysis as mda
>>> import pytim
>>> from pytim.datafiles import *
>>> #
>>> # this is a DPC micelle in water
>>> u = mda.Universe(MICELLE_PDB)
>>> #
>>> # select all lipid molecues
>>> g = u.select_atoms('resname DPC')
In order to calculate the surface atoms, we invoke GITIM, passing
the group g as the group option, and set molecular=False
in order to mark
only the atoms at the surface, and not the whole residues to which these atoms belong.
We override the standard radii (from the gromos 43a1 forcefield) with a simple set that
will match the atom types.
>>> # define radii for atom types
>>> rdict = {'C':1.5, 'O':1.5, 'P':1.6, 'N':1.8}
>>> # calculate the atoms at the interface ...
>>> inter = pytim.GITIM(u, group=g, molecular=False, alpha=2.5, radii_dict=rdict )
>>> #
>>> # ... and write a pdb file (default:'layers.pdb') with layer information in the beta field
>>> inter.writepdb()
The micelle with its surface atoms highlighted in purple looks like this (left: from outside; right: section through the middle)
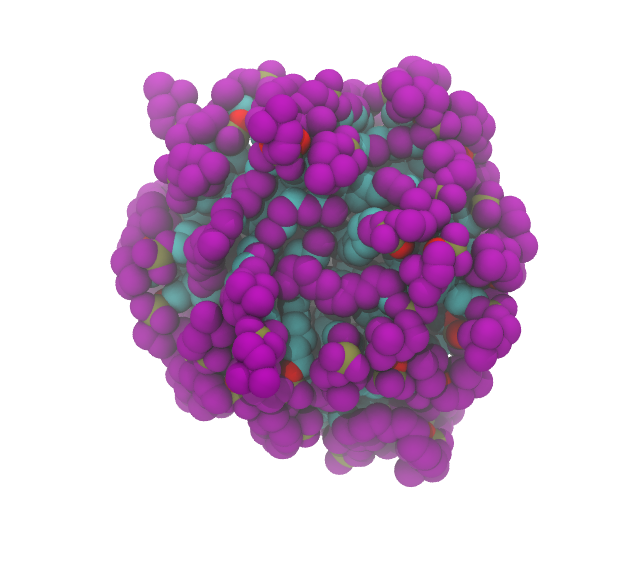
|
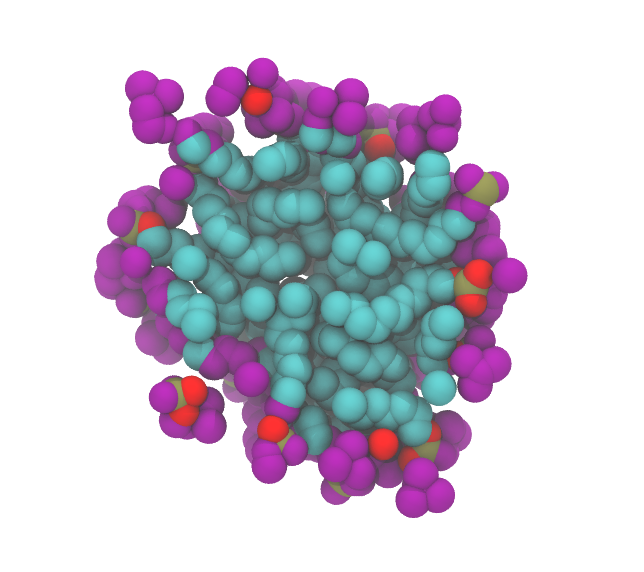
|
Some statistics¶
It’s easy to calculate some simple statistical properties. For example, the percentage of atoms of DPC at the surface is
>>> print ("percentage of atoms at the surface: {:.1f}".format(len(inter.layers[0])*100./len(g)))
percentage of atoms at the surface: 47.2
This is a rather high percentage, but is due to the small size of the micelle (large surface/volume ratio)
We can also easily find out which atom is more likely to be found at the surface:
>>> # we cycle over the names of atoms in the first residue
>>> for name in g.residues[0].atoms.names :
... total = np.sum(g.names==name)
... surface = np.sum(inter.layers[0].names == name )
... print ('{:>4s} ---> {:>2.0f}%'.format(name, surface*100./total))
C1 ---> 97%
C2 ---> 95%
C3 ---> 100%
N4 ---> 98%
C5 ---> 98%
C6 ---> 92%
O7 ---> 75%
P8 ---> 3%
O9 ---> 72%
O10 ---> 83%
O11 ---> 48%
C12 ---> 31%
C13 ---> 29%
C14 ---> 14%
C15 ---> 17%
C16 ---> 15%
C17 ---> 18%
C18 ---> 12%
C19 ---> 18%
C20 ---> 12%
C21 ---> 17%
C22 ---> 17%
C23 ---> 22%
One immediately notices that with this choice of radii, P atoms are rarely at the surface: this is because they are always buried wihin their bonded neighbors in the headgroup. Also, a non negligible part of the fatty tails are also found from time to time at the surface.
